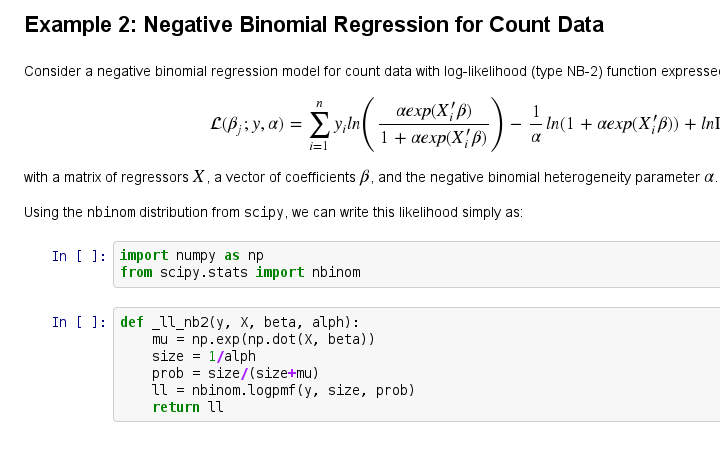
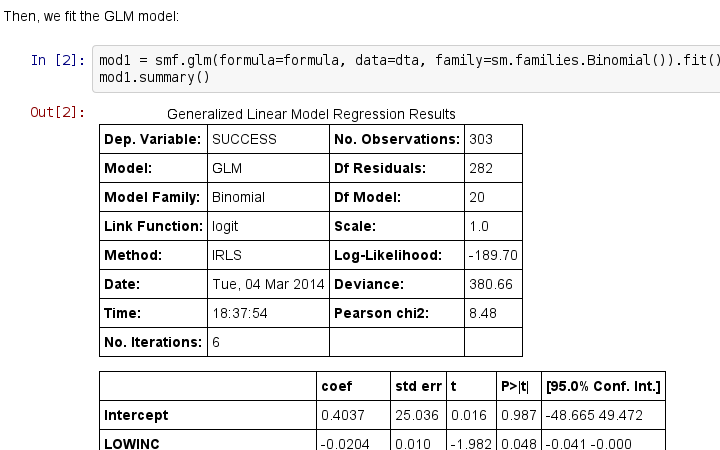
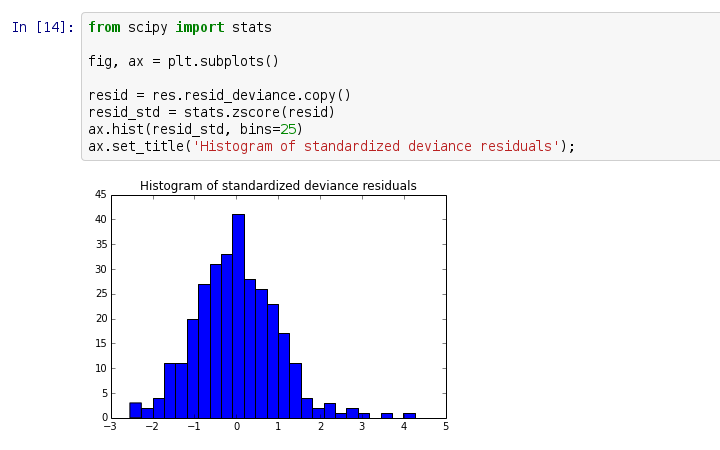
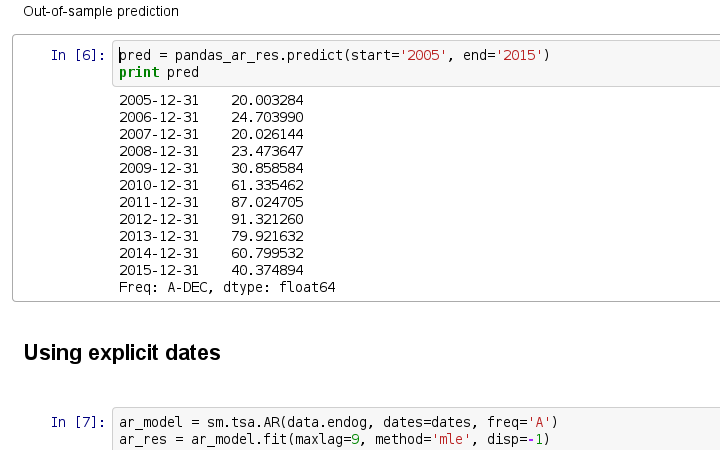
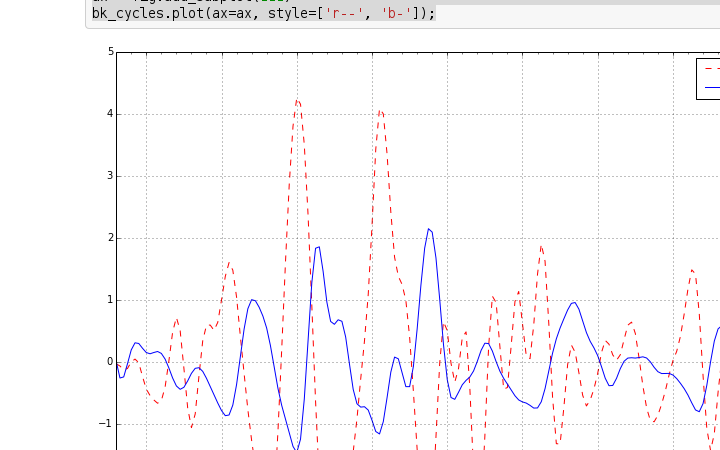
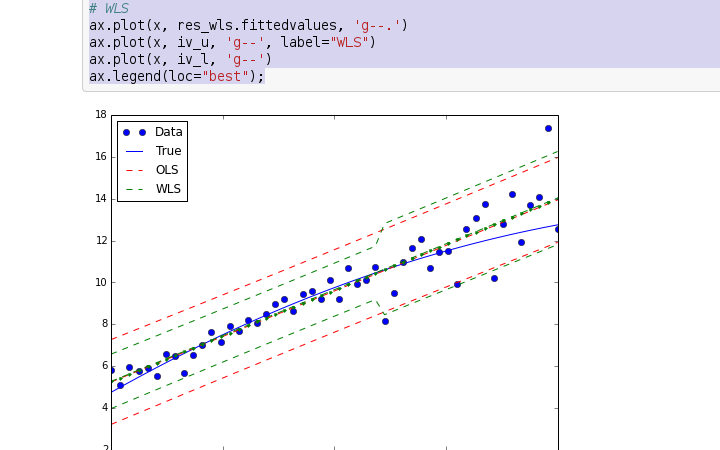
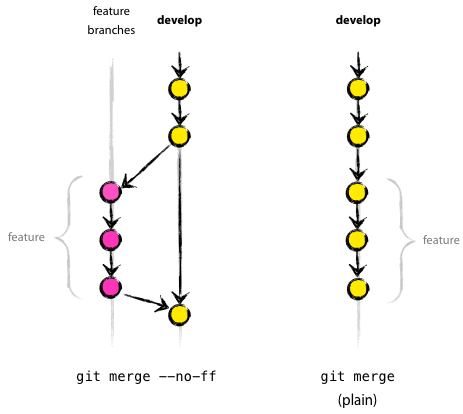
创建项目,准备开始翻译活动
上级
Showing
CHANGES.md
0 → 100644
CONTRIBUTING.rst
0 → 100644
COPYRIGHTS.txt
0 → 100644
INSTALL.txt
0 → 100644
LICENSE.txt
0 → 100644
README.md
0 → 100644
README.rst
0 → 100644
README_l1.txt
0 → 100644
docs/en/GLMNotes.lyx
0 → 100644
此差异已折叠。
docs/en/GLMNotes.pdf
0 → 100644
文件已添加
docs/en/Makefile
0 → 100644
docs/en/README.md
0 → 100644
docs/en/fix_longtable.py
0 → 100644
docs/en/make.bat
0 → 100644
5.4 KB
773 字节
docs/en/source/_static/bullet.gif
0 → 100644
62 字节
168 字节
docs/en/source/_static/facebox.js
0 → 100644
47.3 KB
26.7 KB
18.7 KB
47.3 KB
31.5 KB
41.2 KB
68.0 KB
58.1 KB
37.7 KB
29.3 KB
42.9 KB
34.7 KB
21.9 KB
12.3 KB
29.8 KB
62.0 KB
17.2 KB
124.8 KB
88.3 KB
60.2 KB
32.5 KB
41.7 KB
74.3 KB
31.5 KB
56.2 KB
109.3 KB
23.7 KB
38.8 KB
29.2 KB
25.8 KB
15.1 KB
137.0 KB
107.2 KB
85.5 KB
70.8 KB
123.1 KB
235.4 KB
74.0 KB
194.6 KB
50.1 KB
15.1 KB
38.2 KB
30.7 KB
38.0 KB
36.4 KB
此差异已折叠。
2.7 KB
docs/en/source/_static/minus.gif
0 → 100644
87 字节
docs/en/source/_static/mktree.css
0 → 100644
docs/en/source/_static/mktree.js
0 → 100644
docs/en/source/_static/plus.gif
0 → 100644
89 字节
docs/en/source/_static/scripts.js
0 → 100644
docs/en/source/about.rst
0 → 100644
docs/en/source/anova.rst
0 → 100644
docs/en/source/conf.py
0 → 100644
docs/en/source/contrasts.rst
0 → 100644
docs/en/source/datasets/index.rst
0 → 100644
docs/en/source/dev/examples.rst
0 → 100644
docs/en/source/dev/git_notes.rst
0 → 100644
34.8 KB
docs/en/source/dev/index.rst
0 → 100644
此差异已折叠。
docs/en/source/dev/internal.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/dev/test_notes.rst
0 → 100644
此差异已折叠。
docs/en/source/dev/testing.rst
0 → 100644
docs/en/source/dev/vbench.rst
0 → 100644
此差异已折叠。
docs/en/source/diagnostic.rst
0 → 100644
此差异已折叠。
docs/en/source/discretemod.rst
0 → 100644
此差异已折叠。
docs/en/source/distributions.rst
0 → 100644
此差异已折叠。
docs/en/source/duration.rst
0 → 100644
此差异已折叠。
docs/en/source/emplike.rst
0 → 100644
此差异已折叠。
docs/en/source/endog_exog.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/en/source/examples/README
0 → 100644
此差异已折叠。
docs/en/source/examples/index.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/en/source/extending.rst.TXT
0 → 100644
docs/en/source/faq.rst
0 → 100644
此差异已折叠。
docs/en/source/gee.rst
0 → 100644
此差异已折叠。
docs/en/source/genericmle.rst.TXT
0 → 100644
docs/en/source/gettingstarted.rst
0 → 100644
此差异已折叠。
docs/en/source/glm.rst
0 → 100644
此差异已折叠。
docs/en/source/glm_techn1.rst.TXT
0 → 100644
此差异已折叠。
docs/en/source/glm_techn2.rst.TXT
0 → 100644
此差异已折叠。
docs/en/source/gmm.rst
0 → 100644
此差异已折叠。
docs/en/source/gmm_techn1.rst.TXT
0 → 100644
此差异已折叠。
docs/en/source/graphics.rst
0 → 100644
此差异已折叠。
docs/en/source/images/aw.png
0 → 100644
此差异已折叠。
docs/en/source/images/hl.png
0 → 100644
此差异已折叠。
docs/en/source/images/ht.png
0 → 100644
此差异已折叠。
docs/en/source/images/ls.png
0 → 100644
此差异已折叠。
docs/en/source/images/re.png
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/images/tk.png
0 → 100644
此差异已折叠。
docs/en/source/images/tm.png
0 → 100644
此差异已折叠。
docs/en/source/importpaths.rst
0 → 100644
此差异已折叠。
docs/en/source/imputation.rst
0 → 100644
此差异已折叠。
docs/en/source/index.rst
0 → 100644
此差异已折叠。
docs/en/source/install.rst
0 → 100644
此差异已折叠。
docs/en/source/iolib.rst
0 → 100644
此差异已折叠。
docs/en/source/miscmodels.rst
0 → 100644
此差异已折叠。
docs/en/source/missing.rst
0 → 100644
此差异已折叠。
docs/en/source/mixed_glm.rst
0 → 100644
此差异已折叠。
docs/en/source/mixed_linear.rst
0 → 100644
此差异已折叠。
docs/en/source/multivariate.rst
0 → 100644
此差异已折叠。
docs/en/source/nonparametric.rst
0 → 100644
此差异已折叠。
docs/en/source/pitfalls.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/en/source/plots/bkf_plot.py
0 → 100644
此差异已折叠。
docs/en/source/plots/cff_plot.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/plots/hpf_plot.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/plots/var_plots.py
0 → 100644
此差异已折叠。
docs/en/source/regression.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/release/index.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/en/source/rlm.rst
0 → 100644
此差异已折叠。
docs/en/source/rlm_techn1.rst
0 → 100644
此差异已折叠。
docs/en/source/sandbox.rst
0 → 100644
此差异已折叠。
docs/en/source/statespace.rst
0 → 100644
此差异已折叠。
docs/en/source/stats.rst
0 → 100644
此差异已折叠。
docs/en/source/tools.rst
0 → 100644
此差异已折叠。
docs/en/source/tsa.rst
0 → 100644
此差异已折叠。
docs/en/source/tsastats.rst.TXT
0 → 100644
此差异已折叠。
docs/en/source/vector_ar.rst
0 → 100644
此差异已折叠。
docs/en/sphinxext/LICENSE.txt
0 → 100644
此差异已折叠。
docs/en/sphinxext/MANIFEST.in
0 → 100644
此差异已折叠。
docs/en/sphinxext/README.txt
0 → 100644
此差异已折叠。
docs/en/sphinxext/github.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/GLMNotes.lyx
0 → 100644
此差异已折叠。
docs/zh/GLMNotes.pdf
0 → 100644
此差异已折叠。
docs/zh/Makefile
0 → 100644
此差异已折叠。
docs/zh/README.md
0 → 100644
此差异已折叠。
docs/zh/fix_longtable.py
0 → 100644
此差异已折叠。
docs/zh/make.bat
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/_static/bullet.gif
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/_static/facebox.js
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/_static/minus.gif
0 → 100644
此差异已折叠。
docs/zh/source/_static/mktree.css
0 → 100644
此差异已折叠。
docs/zh/source/_static/mktree.js
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/_static/plus.gif
0 → 100644
此差异已折叠。
docs/zh/source/_static/scripts.js
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/about.rst
0 → 100644
此差异已折叠。
docs/zh/source/anova.rst
0 → 100644
此差异已折叠。
docs/zh/source/conf.py
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/contrasts.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/datasets/index.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/dev/examples.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/dev/git_notes.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/dev/index.rst
0 → 100644
此差异已折叠。
docs/zh/source/dev/internal.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/dev/test_notes.rst
0 → 100644
此差异已折叠。
docs/zh/source/dev/testing.rst
0 → 100644
此差异已折叠。
docs/zh/source/dev/vbench.rst
0 → 100644
此差异已折叠。
docs/zh/source/diagnostic.rst
0 → 100644
此差异已折叠。
docs/zh/source/discretemod.rst
0 → 100644
此差异已折叠。
docs/zh/source/distributions.rst
0 → 100644
此差异已折叠。
docs/zh/source/duration.rst
0 → 100644
此差异已折叠。
docs/zh/source/emplike.rst
0 → 100644
此差异已折叠。
docs/zh/source/endog_exog.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/examples/README
0 → 100644
此差异已折叠。
docs/zh/source/examples/index.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/extending.rst.TXT
0 → 100644
docs/zh/source/faq.rst
0 → 100644
此差异已折叠。
docs/zh/source/gee.rst
0 → 100644
此差异已折叠。
docs/zh/source/genericmle.rst.TXT
0 → 100644
docs/zh/source/gettingstarted.rst
0 → 100644
此差异已折叠。
docs/zh/source/glm.rst
0 → 100644
此差异已折叠。
docs/zh/source/glm_techn1.rst.TXT
0 → 100644
此差异已折叠。
docs/zh/source/glm_techn2.rst.TXT
0 → 100644
此差异已折叠。
docs/zh/source/gmm.rst
0 → 100644
此差异已折叠。
docs/zh/source/gmm_techn1.rst.TXT
0 → 100644
此差异已折叠。
docs/zh/source/graphics.rst
0 → 100644
此差异已折叠。
docs/zh/source/images/aw.png
0 → 100644
此差异已折叠。
docs/zh/source/images/hl.png
0 → 100644
此差异已折叠。
docs/zh/source/images/ht.png
0 → 100644
此差异已折叠。
docs/zh/source/images/ls.png
0 → 100644
此差异已折叠。
docs/zh/source/images/re.png
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/images/tk.png
0 → 100644
此差异已折叠。
docs/zh/source/images/tm.png
0 → 100644
此差异已折叠。
docs/zh/source/importpaths.rst
0 → 100644
此差异已折叠。
docs/zh/source/imputation.rst
0 → 100644
此差异已折叠。
docs/zh/source/index.rst
0 → 100644
此差异已折叠。
docs/zh/source/install.rst
0 → 100644
此差异已折叠。
docs/zh/source/iolib.rst
0 → 100644
此差异已折叠。
docs/zh/source/miscmodels.rst
0 → 100644
此差异已折叠。
docs/zh/source/missing.rst
0 → 100644
此差异已折叠。
docs/zh/source/mixed_glm.rst
0 → 100644
此差异已折叠。
docs/zh/source/mixed_linear.rst
0 → 100644
此差异已折叠。
docs/zh/source/multivariate.rst
0 → 100644
此差异已折叠。
docs/zh/source/nonparametric.rst
0 → 100644
此差异已折叠。
docs/zh/source/pitfalls.rst
0 → 100644
此差异已折叠。
此差异已折叠。
docs/zh/source/plots/bkf_plot.py
0 → 100644
此差异已折叠。
docs/zh/source/plots/cff_plot.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/plots/hpf_plot.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/plots/var_plots.py
0 → 100644
此差异已折叠。
docs/zh/source/regression.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/release/index.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
docs/zh/source/rlm.rst
0 → 100644
此差异已折叠。
docs/zh/source/rlm_techn1.rst
0 → 100644
此差异已折叠。
docs/zh/source/sandbox.rst
0 → 100644
此差异已折叠。
docs/zh/source/statespace.rst
0 → 100644
此差异已折叠。
docs/zh/source/stats.rst
0 → 100644
此差异已折叠。
docs/zh/source/tools.rst
0 → 100644
此差异已折叠。
docs/zh/source/tsa.rst
0 → 100644
此差异已折叠。
docs/zh/source/tsastats.rst.TXT
0 → 100644
此差异已折叠。
docs/zh/source/vector_ar.rst
0 → 100644
此差异已折叠。
docs/zh/sphinxext/LICENSE.txt
0 → 100644
此差异已折叠。
docs/zh/sphinxext/MANIFEST.in
0 → 100644
此差异已折叠。
docs/zh/sphinxext/README.txt
0 → 100644
此差异已折叠。
docs/zh/sphinxext/github.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/incomplete/arima.py
0 → 100644
此差异已折叠。
examples/incomplete/arma2.py
0 → 100644
此差异已折叠。
examples/incomplete/dates.py
0 → 100644
此差异已折叠。
examples/incomplete/glsar.py
0 → 100644
此差异已折叠。
examples/incomplete/ols_table.py
0 → 100644
此差异已折叠。
examples/incomplete/ols_tftest.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/notebooks/formulas.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
examples/notebooks/glm.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/notebooks/gls.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/notebooks/ols.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/notebooks/predict.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/notebooks/wls.ipynb
0 → 100644
此差异已折叠。
此差异已折叠。
examples/python/contrasts.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/python/formulas.py
0 → 100644
此差异已折叠。
examples/python/generic_mle.py
0 → 100644
此差异已折叠。
examples/python/glm.py
0 → 100644
此差异已折叠。
examples/python/glm_formula.py
0 → 100644
此差异已折叠。
examples/python/gls.py
0 → 100644
此差异已折叠。
此差异已折叠。
examples/python/kernel_density.py
0 → 100644
此差异已折叠。
examples/python/ols.py
0 → 100644
此差异已折叠。
examples/python/predict.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
examples/python/tsa_arma_0.py
0 → 100644
此差异已折叠。
examples/python/tsa_arma_1.py
0 → 100644
此差异已折叠。
examples/python/tsa_dates.py
0 → 100644
此差异已折叠。
examples/python/tsa_filters.py
0 → 100644
此差异已折叠。
examples/python/wls.py
0 → 100644
此差异已折叠。
examples/run_all.py
0 → 100644
此差异已折叠。